

Transforming Data with `dplyr`
Adam J Sullivan
Assistant Professor of Biostatistics
Brown University
tidyr
The principles of tidy data provide a standard way to organize data values within a dataset.
--Hadley Wickham (2014)
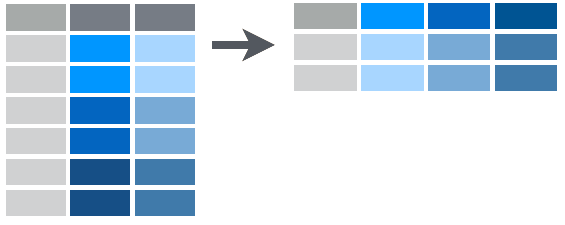
The spread() Function
The spread() Function
- The first
tidyrfunction we will look into is thespread()function. - With
spread()it does similar to what you would expect. - We have a data frame where some of the rows contain information that is really a variable name.
- This means the columns are a combination of variable names as well as some data.
What does it do?
The picture below displays this:
Consider this Data:
## # A tibble: 12 x 4
## country year key value
## <fct> <int> <fct> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583
key column
- Notice that in the column of
key, instead of there being values we see the following variable names:- cases
- population
What Should we see?
- In order to use this data we need to have it so the data frame looks like this instead:
## # A tibble: 6 x 4
## country year cases population
## <fct> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
Using the spread() Function
- Now we can see that we have all the columns representing the variables we are interested in and each of the rows is now a complete observation.
- In order to do this we need to learn about the
spread()function:
The spread() Function
spread(data, key, value)
- Where
datais your dataframe of interest.keyis the column whose values will become variable names.valueis the column where values will fill in under the new variables created fromkey.
Piping
- If we consider piping, we can write this as:
data %>%
spread(key, value)
spread() Example
- Now if we consider table2 , we can see that we have:
## # A tibble: 12 x 4
## country year key value
## <fct> <int> <fct> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583
spread() Example
- Now this table was made for this example so key is the
keyin ourspread()function and value is thevaluein ourspread()function. - We can fix this with the following code:
spread() Example
table2 %>%
spread(key,value)
## # A tibble: 6 x 4
## country year cases population
## <fct> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
spread() Example
- We can now see that we have a variable named
casesand a variable namedpopulation. - This is much more tidy.
On Your Own: RStudio Practice
- In this example we will use the dataset
populationthat is part of tidyverse. - Print this data:
## # A tibble: 1 x 3
## country year population
## <chr> <int> <int>
## 1 Afghanistan 1995 17586073
On Your Own: RStudio Practice
- You should see the table that we have above, now We have a variable named
year, assume that we wish to actually have each year as its own variable. - Using the
spread()function, redo this data so that each year is a variable. - Your data will look like this at the end:
On Your Own: RStudio Practice
## # A tibble: 219 x 20
## country `1995` `1996` `1997` `1998` `1999` `2000` `2001` `2002` `2003`
## <chr> <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 Afghan~ 1.76e7 1.84e7 1.90e7 1.95e7 2.00e7 2.06e7 2.13e7 2.22e7 2.31e7
## 2 Albania 3.36e6 3.34e6 3.33e6 3.33e6 3.32e6 3.30e6 3.29e6 3.26e6 3.24e6
## 3 Algeria 2.93e7 2.98e7 3.03e7 3.08e7 3.13e7 3.17e7 3.22e7 3.26e7 3.30e7
## 4 Americ~ 5.29e4 5.39e4 5.49e4 5.59e4 5.68e4 5.75e4 5.82e4 5.87e4 5.91e4
## 5 Andorra 6.39e4 6.43e4 6.41e4 6.38e4 6.41e4 6.54e4 6.80e4 7.16e4 7.56e4
## 6 Angola 1.21e7 1.25e7 1.28e7 1.31e7 1.35e7 1.39e7 1.44e7 1.49e7 1.54e7
## 7 Anguil~ 9.81e3 1.01e4 1.03e4 1.05e4 1.08e4 1.11e4 1.14e4 1.17e4 1.20e4
## 8 Antigu~ 6.83e4 7.02e4 7.22e4 7.42e4 7.60e4 7.76e4 7.90e4 8.00e4 8.09e4
## 9 Argent~ 3.48e7 3.53e7 3.57e7 3.61e7 3.65e7 3.69e7 3.73e7 3.76e7 3.80e7
## 10 Armenia 3.22e6 3.17e6 3.14e6 3.11e6 3.09e6 3.08e6 3.06e6 3.05e6 3.04e6
## # ... with 209 more rows, and 10 more variables: `2004` <int>,
## # `2005` <int>, `2006` <int>, `2007` <int>, `2008` <int>, `2009` <int>,
## # `2010` <int>, `2011` <int>, `2012` <int>, `2013` <int>
The gather() Function
The gather() Function
- The second
tidyrfunction we will look into is thegather()function. - With
gather()it may not be clear what exactly is going on, but in this case we actually have a lot of column names the represent what we would like to have as data values.
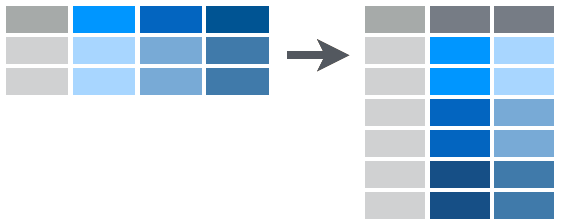
The gather() Function Example
- For example, in the last
spread()practice you created a data frame where variable names were individual years. - This may not be what you want to have so you can use the gather function.
Consider table4:
## # A tibble: 3 x 3
## country `1999` `2000`
## <fct> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766
Table 4
- This looks similar to the table you created in the
spread()practice. - We now wish to change this data frame so that
yearis a variable and 1999 and 2000 become values instead of variables.
In Comes the gather() Function
- We will accomplish this with the gather function:
gather(data, key, value, ...)
- where
datais the data frame you are working with.keyis the name of thekeycolumn to create.valueis the name of thevaluecolumn to create....is a way to specify what columns to gather from.
gather() Example
- In our example here we would do the following:
table4 %>%
gather("year", "cases", 2:3)
## # A tibble: 6 x 3
## country year cases
## <fct> <chr> <int>
## 1 Afghanistan 1999 745
## 2 Brazil 1999 37737
## 3 China 1999 212258
## 4 Afghanistan 2000 2666
## 5 Brazil 2000 80488
## 6 China 2000 213766
gather() Example
- You can see that we have created 2 new columns called
yearandcases. - We filled these with the previous 2nd and 3rd columns.
- Note that we could have done this in many different ways too.
gather() Example
- For example if we knew the years but not which columns we could do this:
table4 %>%
gather("year", "cases", "1999":"2000")
- We could also see that we want to gather all columns except the first so we could have used:
table4 %>%
gather("year", "cases", -1)
On Your Own: RStudio Practice
- Create
population2from last example:
population 2 <- population %>%
spread(year, population)
- Now gather the columns that are labeled by year and create columns
yearandpopulation.
On Your Own: RStudio Practice
In the end your data frame should look like:
## # A tibble: 2 x 3
## country year population
## <chr> <int> <int>
## 1 Afghanistan 1995 17586073
## 2 Afghanistan 1996 18415307
The dplyr Package
The dplyr Package
- Now that we have started to tidy up our data we can see that we have a need to transform this data.
- We may wish to add additional variables.
- The
dplyrpackage allows us to further work with our data.
dplyr Functionality
- With
dplyrwe have five basic verbs that we will learn to work with:filter()select()arrange()mutate()summarize()
Filtering
Filtering
- At this point we will consider how we pick the rows of the data that we wish to work with.
- If you consider many modern data sets, we have so much information that we may not want to bring it all in at once.
- R brings data into the RAM of your computer. This means you can be limited for what size data you can bring in at once.
- Very rarely do you need the entire data set.
- We will focus on how to pick the rows or observations we want now.
Enter the filter() Function
- The
filter()function chooses rows that meet a specific criteria. - We can do this with Base R functions or with
dplyr.
library(dplyr)
Filtering Example
- Let's say that we want to look at the data just for the country of Kenya in 2000
- We could do this without learning a new command and use indexing which we learned yesterday.
gapminder[gapminder$country=="Kenya" & gapminder$year==2002, ]
## # A tibble: 1 x 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Kenya Africa 2002 51.0 31386842 1288.
Filtering Example
- Now this is not very difficult to do, what we have is that we are working with
gapminderand we only want to keep the rows of data therecountry=="Kenyaandyear==2002. - However we could use the
filter()function to do this in a much easier to read format:
filter() Function
filter(.data, ...)
where
.datais a tibble....is a set of arguments the data you want returned needs to meet.
Filtering Example
- This means in our example we could perform the following:
gapminder %>%
filter(country=="Kenya", year==2002)
Finally we could also only do one filtering at a time and chain it:
gapminder %>%
filter(country=="Kenya") %>%
filter(year==2002)
Further Filtering
filter()supports the use of multiple conditions where we can use Boolean.- For example if we wanted to consider only observations that have a life expectancy between 49 and 60, we could do the following:
gapminder %>% filter(lifeExp>=49 & lifeExp<60)
Further Filtering
## # A tibble: 373 x 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Albania Europe 1952 55.2 1282697 1601.
## 2 Albania Europe 1957 59.3 1476505 1942.
## 3 Algeria Africa 1967 51.4 12760499 3247.
## 4 Algeria Africa 1972 54.5 14760787 4183.
## 5 Algeria Africa 1977 58.0 17152804 4910.
## 6 Bahrain Asia 1952 50.9 120447 9867.
## 7 Bahrain Asia 1957 53.8 138655 11636.
## 8 Bahrain Asia 1962 56.9 171863 12753.
## 9 Bahrain Asia 1967 59.9 202182 14805.
## 10 Bangladesh Asia 1982 50.0 93074406 677.
## # ... with 363 more rows
Further Filtering
- We can also use the
filter()function to remove missing data for us. - Previously we learned about a class of functions called
is.foo()where foo represents a data type. - We could choose to only use observations that have life expectancy.
- That means we wish to not have missing data for life expectancy.
gapminder %>% filter(!is.na(lifeExp))
On Your Own: RStudio Practice
Using the filter() function and chaining:
- Choose only rows associated with
- Uganda
- Rwanda
On Your Own: RStudio Practice
Your end result should be:
## # A tibble: 24 x 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Rwanda Africa 1952 40 2534927 493.
## 2 Rwanda Africa 1957 41.5 2822082 540.
## 3 Rwanda Africa 1962 43 3051242 597.
## 4 Rwanda Africa 1967 44.1 3451079 511.
## 5 Rwanda Africa 1972 44.6 3992121 591.
## 6 Rwanda Africa 1977 45 4657072 670.
## 7 Rwanda Africa 1982 46.2 5507565 882.
## 8 Rwanda Africa 1987 44.0 6349365 848.
## 9 Rwanda Africa 1992 23.6 7290203 737.
## 10 Rwanda Africa 1997 36.1 7212583 590.
## # ... with 14 more rows
Selecting
Selecting
- The next logical step would be to select the columns we want as well.
- Many times we have so many columns that we are no interested in for a particular analysis.
- Instead of slowing down your analysis by continuing to run through extra data, we could just select the columns we care about.
Enter the select() Function
- The
select()function chooses columns that we specify. - Again we can do this with base functions or with
dplyr. - We feel that as you continue on with your R usage that you will most likely want to go the route of
dplyrfunctions instead.
Select Example
- Let's say that we want to look at the gapminder data but we are really only interested in the country, life expectancy and year.
- This seems reasonable if we are a customer and wanted to only know these pieces of information. We could do this with indexing :
gapminder[, c("country", "lifeExp", "year")]
select() Function
select(.data, ...)
where
.datais a tibble....are the columns that you wish to have in bare (no quotations)
Selecting Example Continued
We could then do the following
gapminder %>%
select(country, lifeExp, year)
Selecting Example Continued
# A tibble: 1,704 x 3
country lifeExp year
<fctr> <dbl> <int>
1 Afghanistan 28.801 1952
2 Afghanistan 30.332 1957
3 Afghanistan 31.997 1962
4 Afghanistan 34.020 1967
5 Afghanistan 36.088 1972
6 Afghanistan 38.438 1977
7 Afghanistan 39.854 1982
8 Afghanistan 40.822 1987
9 Afghanistan 41.674 1992
10 Afghanistan 41.763 1997
# ... with 1,694 more rows
Removing Columns
- We may wish to pick certain columns that we wish to have but we also may want to remove certain columns.
- It is quite common to de-identify a dataset before actually distributing it to a research team.
- The
select()function will also remove columns.
Removing Columns
- Lets say that we wished to remove the
gdpPercapandpopof the countries:
gapminder %>%
select(-gdpPercap,-pop)
Removing Columns
We also could use a vector for this:
cols <- c("gdpPercap", "pop")
gapminder %>%
select(-one_of(cols))
Removing Columns
- We can also remove columns that contain a certain phrase in the name.
- If we were interested in removing any columns that had to do with time we could search for the phrase "co" in the data and remove them:
gapminder %>%
select(-matches("co"))
Removing Columns
## # A tibble: 1,704 x 4
## year lifeExp pop gdpPercap
## <int> <dbl> <int> <dbl>
## 1 1952 28.8 8425333 779.
## 2 1957 30.3 9240934 821.
## 3 1962 32.0 10267083 853.
## 4 1967 34.0 11537966 836.
## 5 1972 36.1 13079460 740.
## 6 1977 38.4 14880372 786.
## 7 1982 39.9 12881816 978.
## 8 1987 40.8 13867957 852.
## 9 1992 41.7 16317921 649.
## 10 1997 41.8 22227415 635.
## # ... with 1,694 more rows
Unique Observations
- Many times we have a lot of repeats in our data.
- If we just would like to have an account of all things included then we can use the
unique()command. - Lets assume that we wish to know which countries are in the data.
- We do not want to have every country listed over and over again so we ask for unique values:
gapminder %>%
select(country) %>%
unique()
On Your Own: RStudio Practice
- Consider the gapminder data:
gapminder.- Select all but the
popcolumn. - Remove the year and lifeExp from them.
- Select values which contain "p" in them.
- Chain these together so that you run a command and it does all of these things.
- Select all but the
On Your Own: RStudio Practice
Your answer should look like:
## # A tibble: 1,704 x 1
## gdpPercap
## <dbl>
## 1 779.
## 2 821.
## 3 853.
## 4 836.
## 5 740.
## 6 786.
## 7 978.
## 8 852.
## 9 649.
## 10 635.
## # ... with 1,694 more rows
Arranging the Data
Arranging the Data
- We also have need to make sure the data is ordered in a certain manner. This can be easily done in R with the
arrange()function. - Again we can do this in base R but this is not always a clear path.
Arranging the Data Example
- Let's say that we wish to look countries, year and life expectancy.
- Thensay further we want to sort it by Life Expactancy.
- In base R we would have to run the following command:
library(gapminder)
library(tidyverse)
gapminder[order(gapminder$lifeExp), c("country", "year", "lifeExp")]
Enter the arrange() Function
We could do this in an easy manner using the
arrange()function:arrange(.data, ...)Where
.datais a data frame of interest....are the variables you wish to sort by.
Arranging the Data Example Continued
gapminder %>%
select(country,year, lifeExp) %>%
arrange(lifeExp)
Arranging the Data Example Continued
## # A tibble: 1,704 x 3
## country year lifeExp
## <fct> <int> <dbl>
## 1 Rwanda 1992 23.6
## 2 Afghanistan 1952 28.8
## 3 Gambia 1952 30
## 4 Angola 1952 30.0
## 5 Sierra Leone 1952 30.3
## 6 Afghanistan 1957 30.3
## 7 Cambodia 1977 31.2
## 8 Mozambique 1952 31.3
## 9 Sierra Leone 1957 31.6
## 10 Burkina Faso 1952 32.0
## # ... with 1,694 more rows
Arranging the Data Example Continued
- With
arrange()we first useselect()to pick the only columns that we want and then we arrange by thelifeExp. - If we had wished to order them in a descending manner we could have simply used the
desc()function:
gapminder %>%
select(country,year, lifeExp) %>%
arrange(desc(lifeExp))
More Complex Arrange
- Lets consider that we wish to look at the top 3 Life Expectancies for each year.
- Then we wish to order them from largest to smallest Life Expectancy.
- We then need to do the following:
- Group by year.
- Pick the top 3 life expectancy
- order them largest to smallest
More Complex Arrange Continued
gapminder %>%
group_by(year) %>%
top_n(3, lifeExp) %>%
arrange(desc(lifeExp))
- Where
group_by()is a way to group data. This way we perform operations on a group. So top 3 life expectancy are grouped by year.top_n()takes a tibble and returns a specific number of rows based on a chosen value.
More Complex Arrange Continued
## # A tibble: 36 x 6
## # Groups: year [12]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Japan Asia 2002 82 127065841 28605.
## 4 Iceland Europe 2007 81.8 301931 36181.
## 5 Hong Kong, China Asia 2002 81.5 6762476 30209.
## 6 Japan Asia 1997 80.7 125956499 28817.
## 7 Switzerland Europe 2002 80.6 7361757 34481.
## 8 Hong Kong, China Asia 1997 80 6495918 28378.
## 9 Sweden Europe 1997 79.4 8897619 25267.
## 10 Japan Asia 1992 79.4 124329269 26825.
## # ... with 26 more rows
On Your Own: RStudio Practice
- Perform the following operations:
- Group by year.
- use
sample_n()to pick 1 observation per year - Arrange by longest to smallest life expectancy.
On Your Own: RStudio Practice
Your answer may look like:
gapminder %>%
group_by(year) %>%
sample_n(1) %>%
arrange(desc(lifeExp))
Summarizing Data
Summarizing Data
- As you have seen in your own work, being able to summarize information is crucial.
- We need to be able to take out data and summarize it as well.
- We will consider doing this using the
summarise()function.